Publications
Contributions as Book Chapters, Journal Publications, and Proceedings will be listed here as soon preprints are made available online.
Books
Book chapters and full contributions will be listed here as soon as preprints are made available online.
The Hypothesis of Greater Variance, or Why Considering Covert Responses in Human-Computer Interactions
Cutellic P. - in Brain Art, ed. Nijholt A., Springer 2023
Coming soon…
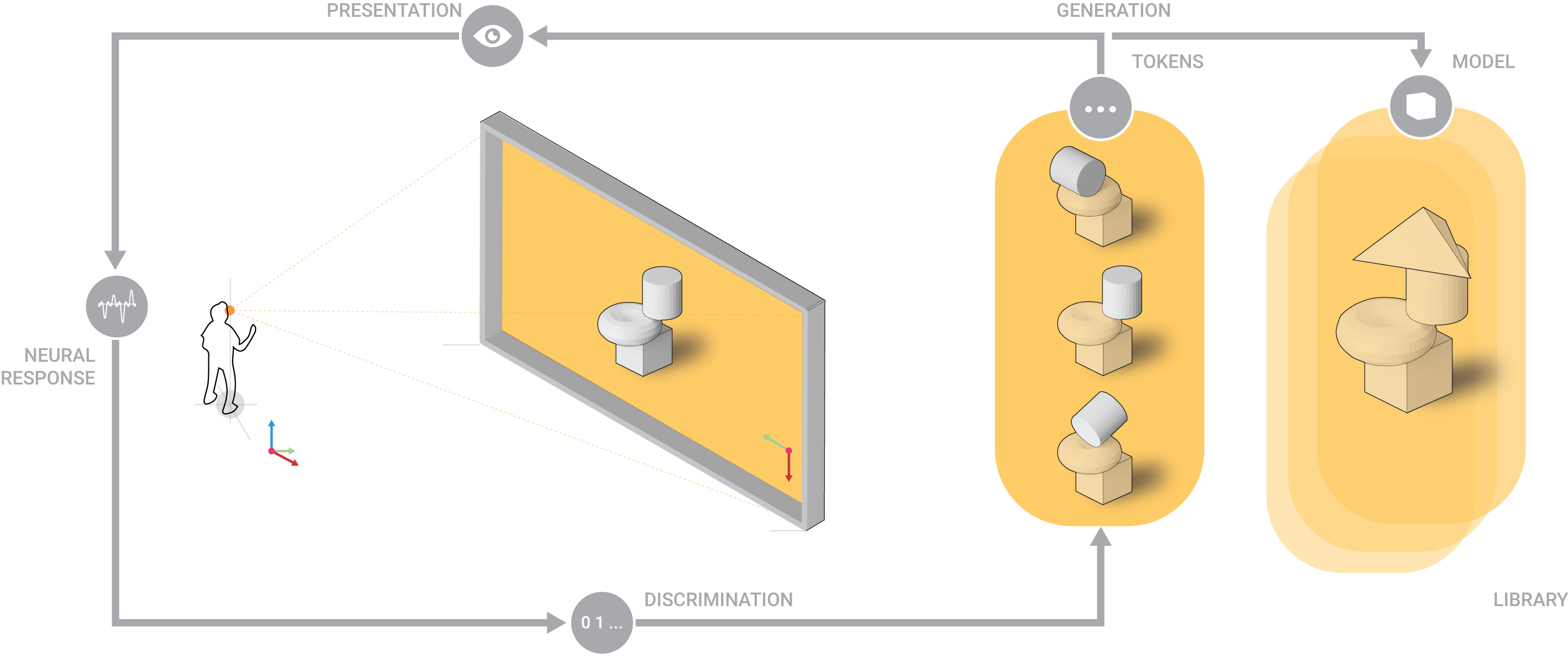
Interactive and Generative modeling loop. Neuramod.
Journals
Journal contributions for work-in-progress and full papers will be listed here as soon as preprints are made available online.
Study Protocol for Randomized Trials on Visual ERP for multiclass discrimination in CAAD applications
Cutellic P., Qureshi N.K. - in BMC Trials, 2022 (Work-In-Progress)
The proposed study aims to observe the detection of Event-Related Potentials’ components and correlated neural phenomena under the visual presentation of complex stimuli and devise processing methods that would generalize their classification for applications in Computer-Aided Architectural Design, where visual complexity becomes an intrinsic feature of the tasks. It aims to investigate the cardinality of discriminative neural patterns correlated with the presentation of complex visual stimuli by detecting subcomponents of these neural phenomena using the designed system on short and prolonged periods of time and involving participants from architecture, visual arts or related fields. Subsequent results will bring to further discussion the role of visual experience in such system and the range it might address in the population segment.
Interactive and Generative modeling loop. Neuramod.
Proceedings
Contributions to Conferences and Symposium Proceedings will be listed here as soon as preprints are made available online.
An Inverse Modeling Method to Estimate Uncertain Spatial Configurations From 2D Information and Time-Based Visual Discriminations
Cutellic P. - in Proceedings of Design Modelling Symposium, Berlin, 2022 (Full Paper)

Interactive and Generative modeling loop. Neuramod.
Growing Shapes with a Generalised Model from Neural Correlates of Visual Discrimination
Cutellic P, Khalid Qureshi N., in Proceedings of the 2020 DigitalFUTURES, Singapore, 2021 (Full Paper)
This paper focuses on the application of visual Event-Related Potentials (ERP) in better generalisations for design and architectural modelling. It makes use of previously built techniques and trained models on EEG signals of a singular individual and observes the robustness of advanced classification models to initiate the development of presentation and classification techniques for enriched visual environments by developing an iterative and generative design process of growing shapes. The pursued interest is to observe if visual ERP as correlates of visual discrimination can hold in structurally similar, but semantically different, experiments and support the discrimination of meaningful design solutions. Following bayesian terms, we will coin this endeavour a Design Belief and elaborate a method to explore and exploit such features decoded from human visual cognition.
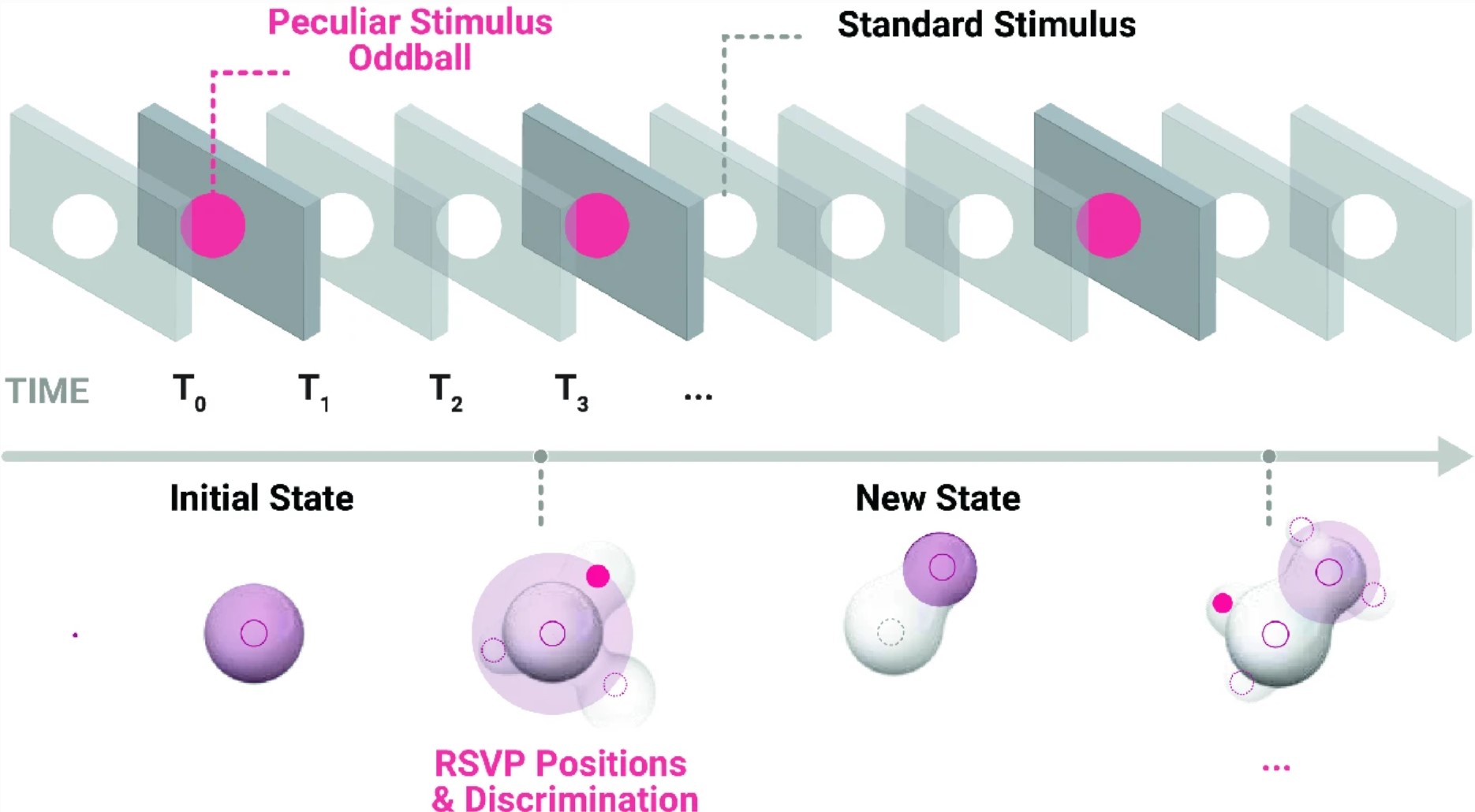
Interactive and Generative modeling loop. Neuramod.
COMING SOON
COMING SOON
COMING SOON